Exploring data warehouse tables with machine learning and Amazon SageMaker notebooks
Are you a data scientist with data warehouse tables that you’d like to explore in your machine learning (ML) environment? If so, read on.
In this post, I show you how to perform exploratory analysis on large datasets stored in your data warehouse and cataloged in your AWS Glue Data Catalog from your Amazon SageMaker notebook. I detail how to identify and explore a dataset in the corporate data warehouse from your Jupyter notebook running on Amazon SageMaker. I demonstrate how to extract the interesting information from Amazon Redshift into Amazon EMR and transform it further there. Then, you can continue analyzing and visualizing your data in your notebook, all in a seamless experience.
| About this blog post | |
| Time to read | 25 minutes |
| Time to complete | 2 hours |
| Cost to complete | Under $10 |
| Learning level | Expert (400) |
| AWS services | Amazon Redshift, Amazon EMR, AWS Glue, Amazon SageMaker |
This post builds on the following prior posts—you may find it helpful to review them first.
- Build Amazon SageMaker notebooks backed by Spark in Amazon EMR
- Access Amazon S3 data managed by AWS Glue Data Catalog from Amazon SageMaker notebooks
Amazon SageMaker overview
Amazon SageMaker is a fully managed ML service. With Amazon SageMaker, data scientists and developers can quickly and easily build and train ML models, and then directly deploy them into a production-ready hosted environment. Amazon SageMaker provides an integrated Jupyter authoring environment for data scientists to perform initial data exploration, analysis, and model building.
The challenge is locating the datasets of interest. If the data is in the data warehouse, you extract the relevant subset of information and load it into your Jupyter notebook for more detailed exploration or modeling. As individual datasets get larger and more numerous, extracting all potentially interesting datasets, loading them into your notebook, and merging them there ceases to be practical and slows productivity. This kind of data combination and exploration can take up to 80% of a data scientist’s time. Increasing productivity here is critical to accelerating the completion of your ML projects.
An increasing number of corporations are using Amazon Redshift as their data warehouse. Amazon Redshift allows you to run complex analytic queries against petabytes of structured data, using sophisticated query optimization, columnar storage on high-performance local disks, and massively parallel query execution. These capabilities make it a magnet for the kind of data that is also of interest to data scientists. However, to perform ML tasks, the data must be extracted into an ML platform so data scientists can operate on it. The capabilities of Amazon Redshift can be used to join and filter the data as needed, then extracting only the relevant data into the ML platform for ML-specific transformation.
Frequently, large corporations also use AWS Glue to manage their data lake. AWS Glue is a fully managed ETL (extract, transform, and load) service. It makes it simple and cost-effective to categorize your data, clean it, enrich it, and move it reliably between various data stores. AWS Glue contains a central metadata repository known as the AWS Glue Data Catalog, which makes the enriched and categorized data in the data lake available for search and querying. You can use the metadata in the Data Catalog to identify the names, locations, content, and characteristics of datasets of interest.
Even after joining and filtering the data in Amazon Redshift, the remaining data may still be too large for your notebook to store and run ML operations on. Operating on extremely large datasets is a task for which Apache Spark on EMR is ideally suited.
Spark is a cluster-computing framework with built-in modules supporting analytics from a variety of languages, including Python, Java, and Scala. Spark on EMR’s ability to scale is a good fit for the large datasets frequently found in corporate data lakes. If the datasets are already defined in your AWS Glue Data Catalog, it becomes easier still to access them, by using the Data Catalog as an external Apache Hive Metastore in EMR. In Spark, you can perform complex transformations that go well beyond the capabilities of SQL. That makes it a good platform for further processing or massaging your data; for example, using the full capabilities of Python and Spark MLlib.
When using the setup described in this post, you use Amazon Redshift to join and filter the source data. Then, you iteratively transform the resulting reduced (but possibly still large) datasets, using EMR for heavyweight processing. You can do this while using your Amazon SageMaker notebook to explore and visualize subsets of the data relevant to the task at hand. The various tasks (joining and filtering; complex transformation; and visualization) have each been farmed out to a service intimately suited to that task.
Solution overview
The first section of the solution walks through querying the AWS Glue Data Catalog to find the database of interest and reviewing the tables and their definitions. The table declarations identify the data location—in this case, Amazon Redshift. The AWS Glue Data Catalog also provides the needed information to build the Amazon Redshift connection string for use in retrieving the data.
The second part of the solution is reading the data into EMR. It applies if the size of the data that you’re extracting from Amazon Redshift is large enough that reading it directly into your notebook is no longer practical. The power of a cluster-compute service, such as EMR, provides the needed scalability.
If the following are true, there is a much simpler solution. For more information, see the Amazon Redshift access demo sample notebook provided with the Amazon SageMaker samples.
- You know the Amazon Redshift cluster that contains the data of interest.
- You know the Amazon Redshift connection information.
- The data you’re extracting and exploring is at a scale amenable to a JDBC connection.
The solution is implemented using four AWS services and some open source components:
- An Amazon SageMaker notebook instance, which provides zero-setup hosted Jupyter notebook IDEs for data exploration, cleaning, and preprocessing. This notebook instance runs:
- Jupyter notebooks
- SparkMagic: A set of tools for interactively working with remote Spark clusters through Livy in Jupyter The SparkMagic project includes a set of magics for interactively running Spark code in multiple languages. Magics are predefined functions that execute supplied commands. The project also includes some kernels that you can use to turn Jupyter into an integrated Spark environment.
- An EMR cluster, running Apache Spark, and:
- Apache Livy: a service that enables easy interaction with Spark on an EMR cluster over a REST interface. Livy enables the use of Spark for interactive web/mobile applications — in this case, from your Jupyter notebook.
- The AWS Glue Data Catalog, which acts as the central metadata repository. Here it’s used as your external Hive Metastore for big data applications running on EMR.
- Amazon Redshift, as your data warehouse.
- The EMR cluster with Spark reads from Amazon Redshift using a Databricks-provided package, Redshift Data Source for Apache Spark.
In this post, all these components interact as shown in the following diagram.
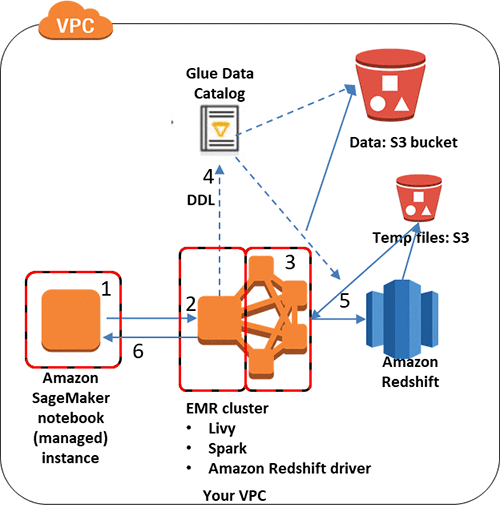
You get access to datasets living on Amazon S3 and defined in the AWS Glue Data Catalog with the following steps:
- You work in your Jupyter SparkMagic notebook in Amazon SageMaker. Within the notebook, you issue commands to the EMR cluster. You can use PySpark commands, or you can use SQL magics to issue HiveQL commands.
- The commands to the EMR cluster are received by Livy, which is running on the cluster.
- Livy passes the commands to Spark, which is also running on the EMR cluster.
- Spark accesses its Hive Metastore to identify the location, DDL, and properties of the cataloged dataset. In this case, the Hive metastore has been set to the Data Catalog.
- You define and run a boto3 function (get_redshift_data, provided below) to retrieve the connection information from the Data Catalog, and issue the command to Amazon Redshift to read the table. The spark-redshift package unloads the table into a temporary S3 file, then loads it into Spark.
- After performing your desired manipulations in Spark, EMR returns the data to your notebook as a dataframe for additional analysis and visualization.
In the sections that follow, you perform these steps on a sample set of tables:
- Use the provided AWS CloudFormation stack to create the Amazon SageMaker notebook instance; EMR cluster with Livy and Spark; and the Amazon Redshift driver. Specify AWS Glue as the cluster’s Hive Metastore; and select an Amazon Redshift cluster. The stack also sets up an AWS Glue connection to the Amazon Redshift cluster, and a crawler to crawl Amazon Redshift.
- Set up some sample data in Amazon Redshift.
- Execute the AWS Glue crawler to access Amazon Redshift and populate metadata about the tables it contains into the Data Catalog.
- From your Jupyter notebook on Amazon SageMaker:
- Use the Data Catalog information to locate the tables of interest, and extract the connection information for Amazon Redshift.
- Read the tables from Amazon Redshift, pulling the data into Spark. You can filter or aggregate the Amazon Redshift data as needed during the unload operation.
- Further transform the data in Spark, transforming it into the desired output.
- Pull the reduced dataset into your notebook, and perform some rudimentary ML on it.
Set up the solution infrastructure
First, you launch a predefined AWS CloudFormation stack to set up the infrastructure components. The AWS CloudFormation stack sets up the following resources:
- An EMR cluster with Livy and Spark, using the AWS Glue Data Catalog as the external Hive compatible Metastore. In addition, it configures Livy to use the same Metastore as the EMR cluster.
- An S3 bucket.
- An Amazon SageMaker notebook instance, along with associated components:
- An IAM role for use by the notebook instance. The IAM role has the managed role AmazonSageMakerFullAccess, plus access to the S3 bucket created above.
- A security group, used for the notebook instance.
- An Amazon SageMaker lifecycle configuration that configures Livy to access the EMR cluster launched by the stack, and copies in a predefined Jupyter notebook with the sample code.
- An Amazon Redshift cluster, in its own security group. Ports are opened to allow EMR, Amazon SageMaker, and the AWS Glue crawler to access it.
- An AWS Glue database, an AWS Glue connection specifying the Amazon Redshift cluster as the target, and an AWS Glue crawler to crawl the connection.
To see this solution in operation in us-west-2, launch the stack from the following button. The total solution costs around $1.00 per hour to run. Remember to delete the AWS CloudFormation stack when you’ve finished with the solution to avoid additional charges.
- Choose Launch Stack and choose Next.
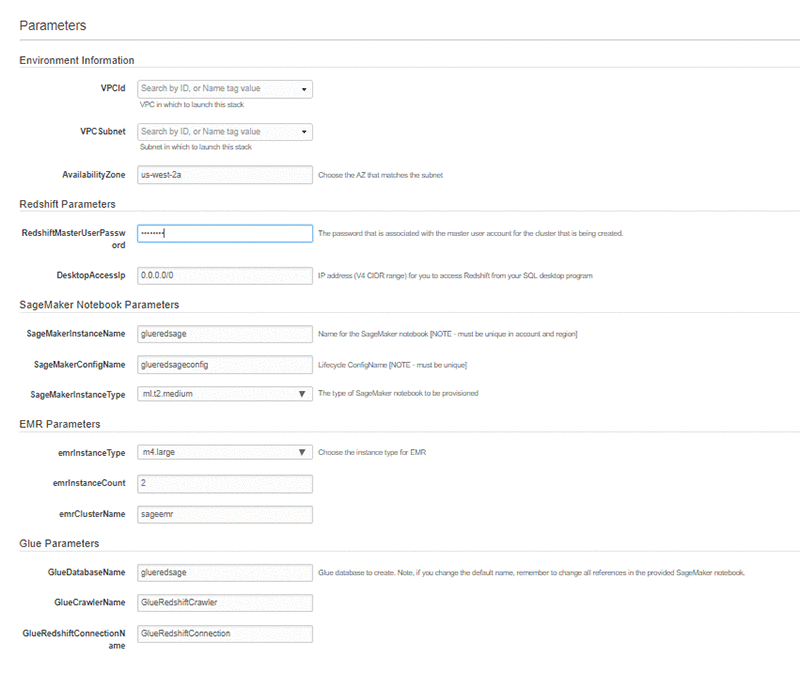
- Update the following parameters for your environment:
- Amazon Redshift password—Must contain at least one uppercase letter, one lowercase letter, and one number.
- VPCId—Must have internet access and an Amazon S3 VPC endpoint. You can use the default VPC created in your account. In the Amazon VPC dashboard, choose Endpoints. Check that the chosen VPC has the following endpoint: com.amazonaws.us-west-2.s3. If not, create one.
- VPCSubnet—Must have internet access, to allow needed software components to be installed.
- Availability Zone—Must match the chosen subnet.
The Availability Zone information and S3 VPC endpoint are used by the AWS Glue crawler to access Amazon Redshift.
- Leave the default values for the other parameters. Changing the AWS Glue database name requires changes to the Amazon SageMaker notebook that you run in a later step. The following screenshot shows the default parameters.

- Choose Next.
- Select I acknowledge that AWS CloudFormation might create IAM resources with custom names, and I acknowledge that AWS CloudFormation might require the following capability: CAPABILITY_AUTO_EXPAND.
- Choose Create.
Wait for the AWS CloudFormation master stack and its nested stacks to reach a status of CREATE_COMPLETE. It can take up to 45 minutes to deploy.
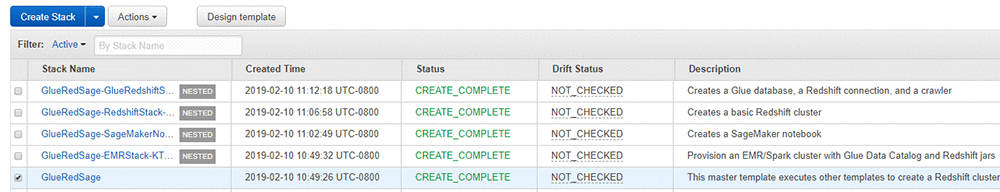
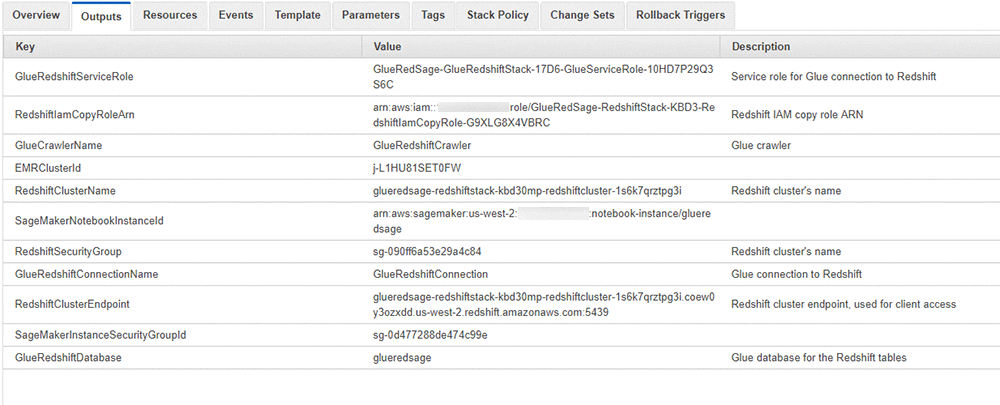
On the master stack, check the Outputs tab for the resources created. You use the key-value data in the next steps. The following screenshot shows the resources that I created but your values will differ.
Add sample data to Amazon Redshift
Using the RedshiftClusterEndpoint from your CloudFormation outputs, the master user name (masteruser), the password you specified in the AWS CloudFormation stack, and the Redshift database of ‘dev’, connect to your Amazon Redshift cluster using your favorite SQL client. Use one of the following methods:
- The Query Editor on the Amazon Redshift console, as described in Querying a Database Using the Query Editor
- A SQL workbench, such as Aginity Workbench for Amazon Redshift
- SQL Workbench/J
The sample data to use comes from Step 6: Load Sample Data from Amazon S3. This data contains the ticket sales for events in several categories, along with information about the categories “liked” by the purchasers. Later, you use this data to calculate the correlation between liking a category and attending events (and then further exploration as desired).
Run the table creation commands followed by the COPY commands. Insert the RedshiftIamCopyRoleArn IAM role created by AWS CloudFormation in the COPY commands. At the end of this sequence, the sample data is in Amazon Redshift, in the public schema. Explore the data in the table, using SQL. You explore the same data again in later steps. You now have an Amazon Redshift data warehouse with several normalized tables containing data related to event ticket sales.
Try the following query. Later, you use this same query (minus the limit) from Amazon SageMaker to retrieve data from Amazon Redshift into EMR and Spark. It also helps confirm that you’ve loaded the data into Amazon Redshift correctly.
The ‘like’ fields contain nulls. Convert these to ‘false’ here, to simplify later processing.
Use an AWS Glue crawler to add tables to the Data Catalog
Now that there’s sample data in the Amazon Redshift cluster, the next step is to make the Amazon Redshift tables visible in the AWS Glue Data Catalog. The AWS CloudFormation template set up the components for you: an AWS Glue database, a connection to Amazon Redshift, and a crawler. Now you run the crawler, which reads Amazon Redshift’s catalog and populate the Data Catalog with that information.
First, test that the AWS Glue connection can connect to Amazon Redshift:
- In the AWS Glue console, in the left navigation pane, choose Connections.
- Select the connection GlueRedshiftConnection, and choose Test Connection.
- When asked for an IAM role, choose the GlueRedshiftService role created by the AWS CloudFormation template.
- Wait while AWS Glue tests the connection. If it successfully does so, you see the message GlueRedshiftConnection connected successfully to your instance. If it does not, the most likely cause is that the subnet, VPC, and Availability Zone did not match. Or, it could be that the subnet is missing an S3 endpoint or internet access.
Next, retrieve metadata from Amazon Redshift about the tables that exist in the Amazon Redshift database noted in the AWS CloudFormation template parameters. To do so, run the AWS Glue crawler that the AWS CloudFormation template created:
- In the AWS Glue console, choose Crawlers in the left-hand navigation bar.
- Select GlueRedshiftCrawler in the crawler list, and choose Run Crawler. If asked for an IAM role, choose the GlueRedshiftService role created by the AWS CloudFormation template.
- Wait as the crawler runs. It should complete in two or three minutes. You see the status change to Starting, then Running, Stopping, and finally Ready.
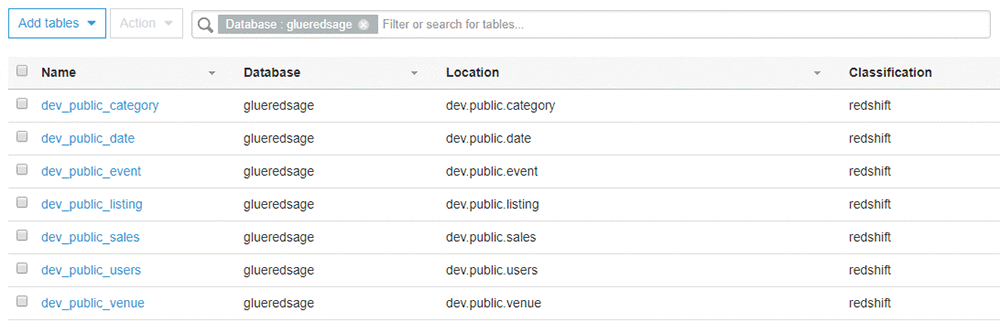
- When the crawler status is Ready, check the column under Tables Added. You should see that seven tables have been added.
To review the tables the crawler added, use the following steps:
- Choose Databases and select the database named glueredsage. This database was created by the AWS CloudFormation stack.
- Choose Tables in glueredsage.
You should see the tables that you created in Amazon Redshift listed, as shown in the screenshot that follows. The AWS Glue table name is made up of the database (dev), the schema (public), and the actual table name from Amazon Redshift (for example, date). The AWS Glue classification is Amazon Redshift.
You access this metadata from your Jupyter notebook in the next step.
Access data defined in AWS Glue Data Catalog from the notebook
In this section, you locate the Amazon Redshift data of interest in the AWS Glue Data Catalog and get the data from Amazon Redshift, from an Amazon SageMaker notebook.
- In the Amazon SageMaker console, in the left navigation pane, choose Notebook instances.
- Next to the notebook started by your AWS CloudFormation stack, choose Open Jupyter.
You see a page similar to the screenshot that follows. The Amazon SageMaker lifecycle configuration in the CF stack automatically uploaded the notebook Using_SageMaker_Notebooks_to_access_Redshift_via_Glue.ipynb to your Jupyter dashboard.
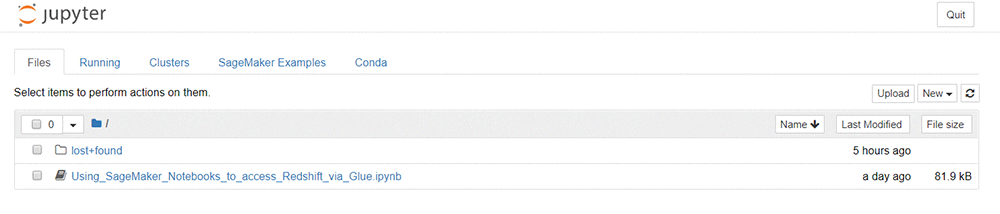
Open the notebook. The kernel type is “SparkMagic (PySpark)”. Alternatively, you can browse the static results of a prior run in HTML format. The following links take you to the relevant section in this version.
Begin executing the cells in the notebook, following the instructions there. The instructions there walk you through:
- Accessing the Spark cluster from your local notebook via Livy, and issuing a simple Pyspark statement from your local notebook to show how you can use Pyspark in this environment.
- Listing the databases in your AWS Glue Data Catalog, and showing the tables in the AWS Glue database, glueredsage, that you set up previously via the AWS CloudFormation template.Here, you use a couple of Python helper functions to access the Data Catalog from your local notebook. You can identify the tables of interest from the Data Catalog, and see that they’re stored in Amazon Redshift. This is your clue that you must connect to Amazon Redshift to read this data.
- Retrieving Amazon Redshift connection information from the Data Catalog for the tables of interest.
- Retrieving the data relevant to your planned research problem from a series of Amazon Redshift tables into Spark EMR using two methods: retrieving the full table, or, by executing a SQL that joins and filters the data.First, you retrieve a small Amazon Redshift table containing some metadata — the categories of events. Then, you perform a complex query that pulls back a flattened dataset containing data about which eventgoers in which cities like what types of events (Broadway, Jazz, classical, etc.). Irrelevant data is not retrieved for further analysis. The data comes back as a Spark data frame, on which you can perform additional analysis.
- Using the resulting (potentially large) dataframe on EMR to first perform some ML functions in Spark: converting several columns into one-hot vector representations and calculating correlations between them. The dataframe of correlations is much smaller, and is practical to process on your local notebook.
- Lastly, working with the processed data frame in your local notebook instance. Here you visualize the (much smaller) results of your correlations locally.
Here’s the result of your initial analysis, showing the correlation between event attendance versus categories of events liked:
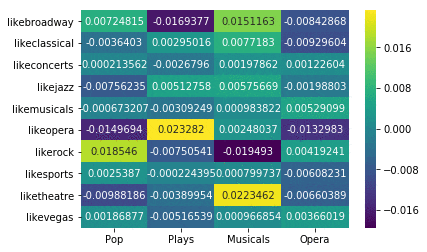
You can see that, based on these ticket purchases and event attendances, the likes and event categories are only weakly correlated (max correlation is 0.02). Though the correlations are weak, relatively speaking:
- Liking theatre is positively correlated with attending musicals.
- Liking opera is positively correlated with attending plays.
- Liking rock is negatively correlated with attending musicals.
- Liking Broadway is negatively correlated with attending plays (surprisingly!).
Debugging your connection
If your notebook does not connect to your EMR cluster, review the following information to see where the problem lies.
Amazon SageMaker notebook instances can use a lifecycle configuration. With a lifecycle configuration, you can provide a Bash script to be run whenever an Amazon SageMaker notebook instance is created, or when it is restarted after having been stopped. The AWS CloudFormation template uses a creation-time script to configure the Livy configuration on the notebook instance with the address of the EMR master instance created earlier. The most common sources of connection difficulties are as follows:
- Not having the correct settings in livy.conf.
- Not having the correct ports open on the security groups between the EMR cluster and the notebook instance.
When the notebook instance is created or started, the results of running the lifecycle config are captured in an Amazon CloudWatch Logs log group called /aws/sagemaker/NotebookInstances. This log group has a stream for
Next, check the Livy configuration and EMR access on the notebook instance. In the Jupyter files dashboard, choose New, Terminal. This opens a shell for you on the notebook instance.
The Livy config file is stored in: /home/ec2-user/SageMaker/.sparkmagic/config.json. Check to make sure that your EMR cluster IP address has replaced the original http://localhost:8998 address in three places in the file.
If you are receiving errors during data retrieval from Amazon Redshift, check whether the request is getting to Amazon Redshift.
- In the Amazon Redshift console, choose Clusters and select the cluster started by the AWS CloudFormation template.
- Choose Queries.
- Your request should be in the list of the SQL queries that the Amazon Redshift cluster has executed. If it isn’t, check that the connection to Amazon Redshift is working, and you’ve used the correct IAM copy role, userid, and password.
A last place to check is the temporary S3 directory that you specified in the copy statement. You should see a folder placed there with the data that was unloaded from Amazon Redshift.
Extending the solution and using in production
The example provided uses a simple dataset and SQL to allow you to more easily focus on the connections between the components. However, the real power of the solution comes from accessing the full capabilities of your Amazon Redshift data warehouse and the data within. You can use far more complex SQL queries—with joins, aggregations, and filters—to manipulate, transform, and reduce the data within Amazon Redshift. Then, pull back the subsets of interest into Amazon SageMaker for more detailed exploration.
This section touches on three additional questions:
- What about merging Amazon Redshift data with data in S3?
- What about moving from the data-exploration phase into training your ML model, and then to production?
- How do you replicate this solution?
Using Redshift Spectrum in this solution
During this data-exploration phase, you may find that some additional data exists on S3 that is useful in combination with the data housed on Amazon Redshift. It’s straightforward to merge the two, using the power of Amazon Redshift Spectrum. Amazon Redshift Spectrum directly queries data in S3, using the same SQL syntax of Amazon Redshift. You can also run queries that span both the frequently accessed data stored locally in Amazon Redshift and your full datasets stored cost-effectively in S3.
To use this capability in from your Amazon SageMaker notebook:
- First, follow the instructions for Cataloging Tables with a Crawler to add your S3 datasets to your AWS Glue Data Catalog.
- Then, follow the instructions in Creating External Schemas for Amazon Redshift Spectrum to add an existing external schema to Amazon Redshift. You need the permissions described in Policies to Grant Minimum Permissions.
After the external schema is defined in Amazon Redshift, you can use SQL to read the S3 files from Amazon Redshift. You can also seamlessly join, aggregate, and filter the S3 files with Amazon Redshift tables.
In exactly the same way, you can use SQL from within the notebook to read the combined S3 and Amazon Redshift data into Spark/EMR. From there, read it into your notebook, using the functions already defined.
Moving from exploration to training and production
The pipeline described here—reading directly from Amazon Redshift—is optimized for the data-exploration phase of your ML project. During this phase, you’re likely iterating quickly across different datasets, seeing which data and which combinations are useful for the problem you’re solving.
After you’ve settled on the data to be used for training, it is more appropriate to materialize the final SQL into an extract on S3. The dataset on S3 can then be used for the training phase, as is demonstrated in the sample Amazon SageMaker notebooks.
Deployment into production has different requirements, with a different data access pattern. For example, the interactive responses needed by online transactions are not a good fit for Amazon Redshift. Consider the needs of your application and data pipeline, and engineer an appropriate combination of data sources and access methods for that need. Replicating this solution
Cleanup
To avoid additional charges, remember to delete the AWS CloudFormation stack when you’ve finished with the solution.
Conclusion
By now, you can see the true power of this combination in exploring data that’s in your data lake and data warehouse:
- Expose data via the AWS Glue Data Catalog.
- Use the scalability and processing capabilities of Amazon Redshift and Amazon EMR to preprocess, filter, join, and aggregate data from your Amazon S3 data lake data.
- Your data scientists can use tools they’re familiar with—Amazon SageMaker, Jupyter notebooks, and SQL—to quickly explore and visualize data that’s already been cataloged.
Another source of friction has been removed, and your data scientists can move at the pace of business.
About the Author
 Veronika Megler is a Principal Consultant, Big Data, Analytics & Data Science, for AWS Professional Services. She holds a PhD in Computer Science, with a focus on spatio-temporal data search. She specializes in technology adoption, helping customers use new technologies to solve new problems and to solve old problems more efficiently and effectively.
Veronika Megler is a Principal Consultant, Big Data, Analytics & Data Science, for AWS Professional Services. She holds a PhD in Computer Science, with a focus on spatio-temporal data search. She specializes in technology adoption, helping customers use new technologies to solve new problems and to solve old problems more efficiently and effectively.
Tags: Archive

Leave a Reply