Securing data analytics with an Amazon SageMaker notebook instance and Kerberized Amazon EMR cluster
Ever since Amazon SageMaker was introduced at AWS re:Invent 2017, customers have used the service to quickly and easily build and train machine learning (ML) models and directly deploy them into a production-ready hosted environment. SageMaker notebook instances provide a powerful, integrated Jupyter notebook interface for easy access to data sources for exploration and analysis. You can enhance the SageMaker capabilities by connecting the notebook instance to an Apache Spark cluster running on Amazon EMR. It gives data scientists and engineers a common instance with shared experience where they can collaborate on AI/ML and data analytics tasks.
If you’re using a SageMaker notebook instance, you may need a way to allow different personas (such as data scientists and engineers) to do different tasks on Amazon EMR with a secure authentication mechanism. For example, you might use the Jupyter notebook environment to build pipelines in Amazon EMR to transform datasets in the data lake, and later switch personas and use the Jupyter notebook environment to query the prepared data and perform advanced analytics on it. Each of these personas and actions may require their own distinct set of permissions to the data.
To address this requirement, you can deploy a Kerberized EMR cluster. Amazon EMR release version 5.10.0 and later supports MIT Kerberos, which is a network authentication protocol created by the Massachusetts Institute of Technology (MIT). Kerberos uses secret-key cryptography to provide strong authentication so passwords or other credentials aren’t sent over the network in an unencrypted format.
This post walks you through connecting a SageMaker notebook instance to a Kerberized EMR cluster using SparkMagic and Apache Livy. Users are authenticated with Kerberos Key Distribution Center (KDC), where they obtain temporary tokens to impersonate themselves as different personas before interacting with the EMR cluster with appropriately assigned privileges.
This post also demonstrates how a Jupyter notebook uses PySpark to download the COVID-19 database in CSV format from the Johns Hopkins GitHub repository. The data is transformed and processed by Pandas and saved to an S3 bucket in columnar Parquet format referenced by an Apache Hive external table hosted on Amazon EMR.
Solution walkthrough
The following diagram depicts the overall architecture of the proposed solution. A VPC with two subnets are created: one public, one private. For security reasons, a Kerberized EMR cluster is created inside the private subnet. It needs access to the internet to access data from the public GitHub repo, so a NAT gateway is attached to the public subnet to allow for internet access.
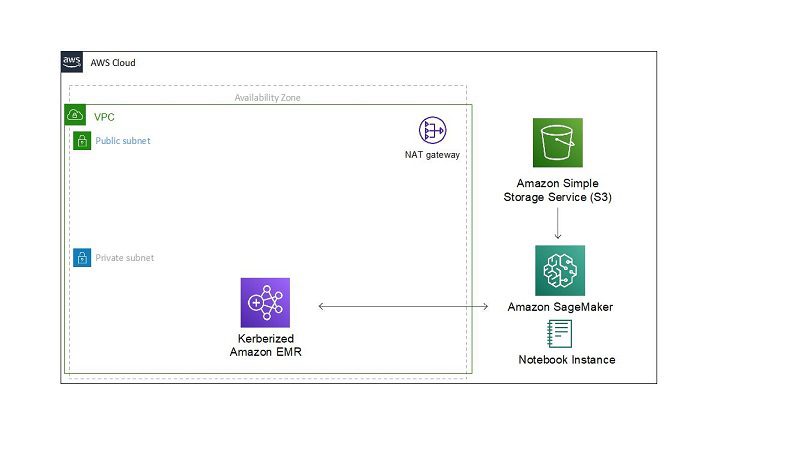
The Kerberized EMR cluster is configured with a bootstrap action in which three Linux users are created and Python libraries are installed (Pandas, requests, and Matplotlib).
You can set up Kerberos authentication a few different ways (for more information, see Kerberos Architecture Options):
- Cluster dedicated KDC
- Cluster dedicated KDC with Active Directory cross-realm trust
- External KDC
- External KDC integrated with Active Directory
The KDC can have its own user database or it can use cross-realm trust with an Active Directory that holds the identity store. For this post, we use a cluster dedicated KDC that holds its own user database. First, the EMR cluster has security configuration enabled to support Kerberos and is launched with a bootstrap action to create Linux users on all nodes and install the necessary libraries. A bash step is launched right after the cluster is ready to create HDFS directories for the Linux users with default credentials that are forced to change as soon as the users log in to the EMR cluster for the first time.
A SageMaker notebook instance is spun up, which comes with SparkMagic support. The Kerberos client library is installed and the Livy host endpoint is configured to allow for the connection between the notebook instance and the EMR cluster. This is done through configuring the SageMaker notebook instance’s lifecycle configuration feature. We provide sample scripts later in this post to illustrate this process.
Fine-grained user access control for EMR File System
The EMR File System (EMRFS) is an implementation of HDFS that all EMR clusters use for reading and writing regular files from Amazon EMR directly to Amazon Simple Storage Service (Amazon S3). The Amazon EMR security configuration enables you to specify the AWS Identity and Access Management (IAM) role to assume when a user or group uses EMRFS to access Amazon S3. Choosing the IAM role for each user or group enables fine-grained access control for EMRFS on multi-tenant EMR clusters. This allows different personas to be associated with different IAM roles to access Amazon S3.r
Deploying the resources with AWS CloudFormation
You can use the provided AWS CloudFormation template to set up this architecture’s building blocks, including the EMR cluster, SageMaker notebook instance, and other required resources. The template has been successfully tested in the us-east-1 Region.
Complete the following steps to deploy the environment:
- Sign in to the AWS Management Console as an IAM power user, preferably an admin user.
- Choose Launch Stack to launch the CloudFormation template:
- Choose Next.
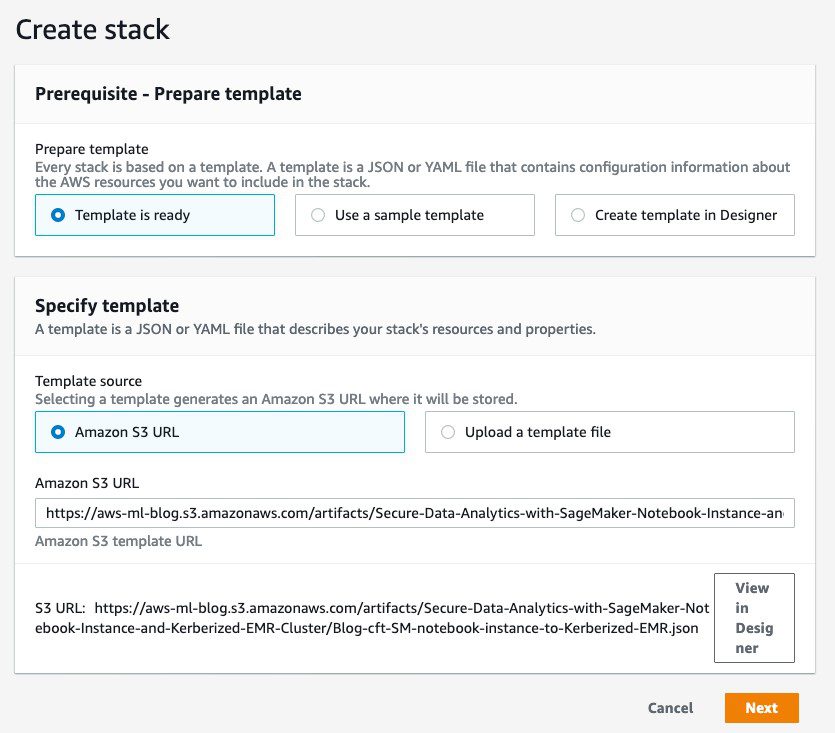
- For Stack name, enter a name for the stack (for example,
blog). - Leave the other values as default.
- Continue to choose Next and leave other parameters at their default.
- On the review page, select I acknowledge that AWS CloudFormation might create IAM resources with custom names.
- Choose Create stack.
Wait until the status of the stack changes from CREATE_IN_PROGRESS to CREATE_COMPLETE. The process usually takes about 10–15 minutes.
After the environment is complete, we can investigate what the template provisioned.
Notebook instance lifecycle configuration
Lifecycle configurations perform the following tasks to ensure a successful Kerberized authentication between the notebook and EMR cluster:
- Configure the Kerberos client on the notebook instance
- Configure SparkMagic to use Kerberos authentication
You can view your provisioned lifecycle configuration, SageEMRConfig, on the Lifecycle configuration page on the SageMaker console.
The template provisioned two scripts to start and create your notebook: start.sh and create.sh, respectively. The scripts replace {EMRDNSName} with your own EMR cluster primary node’s DNS hostname during the CloudFormation deployment.
EMR cluster security configuration
Security configurations in Amazon EMR are templates for different security setups. You can create a security configuration to conveniently reuse a security setup whenever you create a cluster. For more information, see Use Security Configurations to Set Up Cluster Security.
To view the EMR security configuration created, complete the following steps:
- On the Amazon EMR console, choose Security configurations.
- Expand the security configuration created. Its name begins with
blog-securityConfiguration.
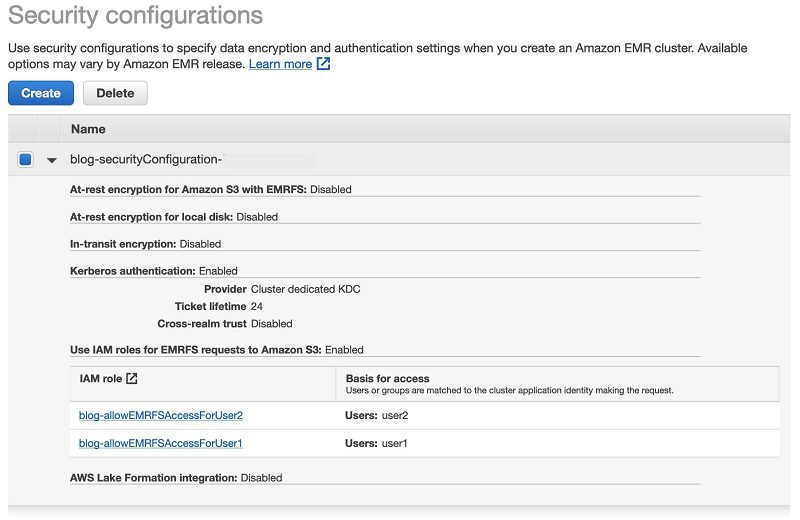
Two IAM roles are created as part of the solution in the CloudFormation template, one for each EMR user (user1 and user2). The two users are created during the Amazon EMR bootstrap action.
- Choose the role for user1,
blog-allowEMRFSAccessForUser1.
The IAM console opens and shows the summary for the IAM role.
- Expand the policy attached to the role
blog-emrFS-user1.
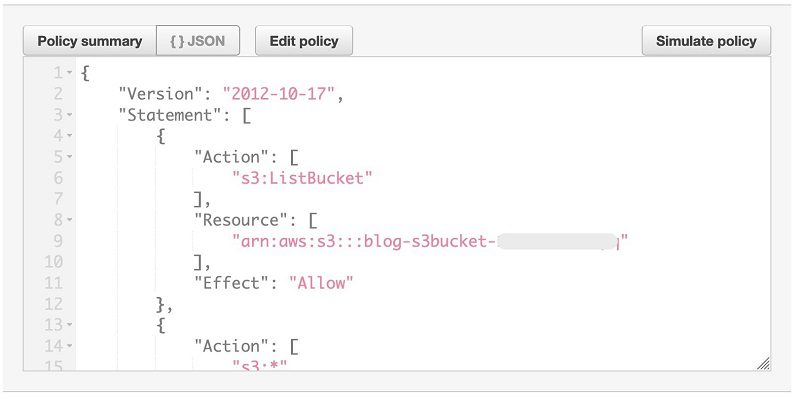
This is the S3 bucket the CloudFormation template created to store the COVID-19 datasets.
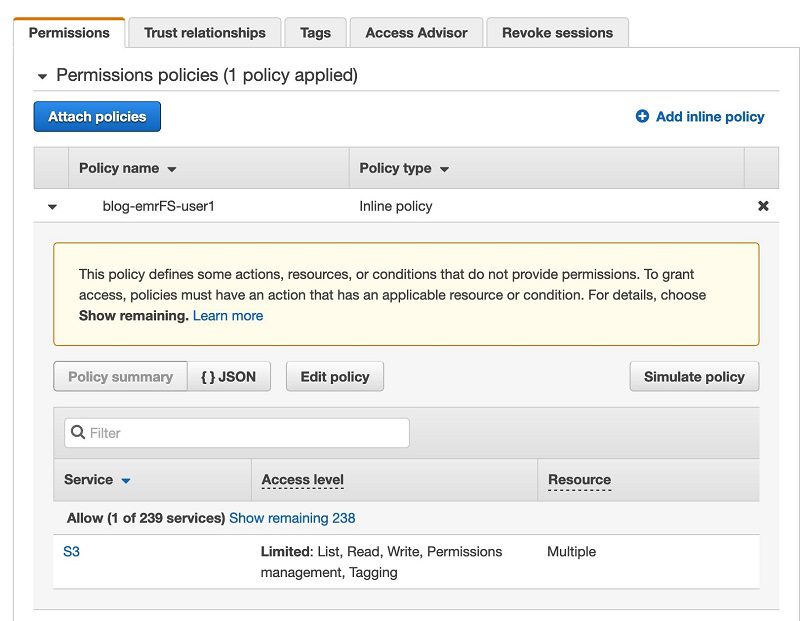
- Choose {} JSON.
You can see the policy definition and permissions to the bucket named blog-s3bucket-xxxxx.
- Return to the EMR security configuration.
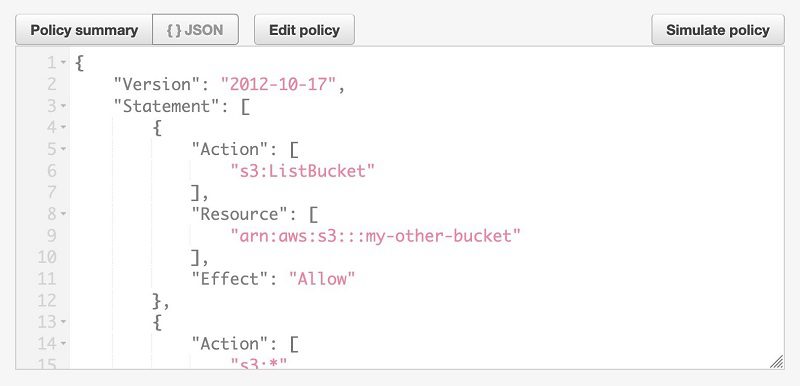
- Choose the IAM policy for user2,
blog-allowEMRFSAccessForUser2. - Expand the policy attached to the role,
blog-emrFS-user2. - Choose {} JSON.
You can see the policy definition and permissions to the bucket named my-other-bucket.
Authenticating with Kerberos
To use these IAM roles, you authenticate via Kerberos from the notebook instance to the EMR cluster KDC. The authenticated user inherits the permissions associated with the policy of the IAM role defined in the Amazon EMR security configuration.
To authenticate with Kerberos in the SageMaker notebook instance, complete the following steps:
- On the SageMaker console, under Notebook, choose Notebook instances.
- Locate the instance named
SageEMR. - Choose Open JupyterLab.
- On the File menu, choose New.
- Choose Terminal.
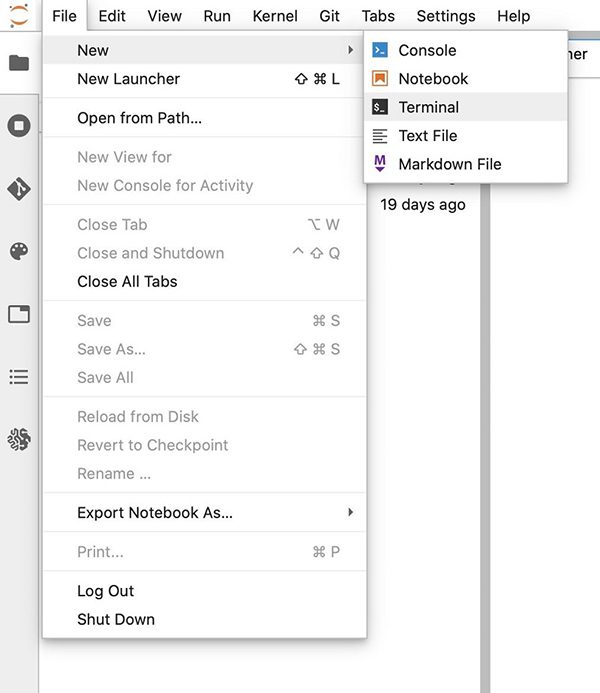
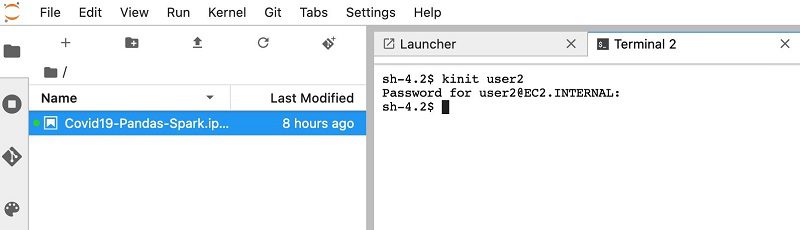
- Enter
kinitfollowed by the usernameuser2. - Enter the user’s password.
The initial default password is pwd2. The first time logging in, you’re prompted to change the password.
- Enter a new password.
- Enter
klistto view the Kerbereos ticket for user2.
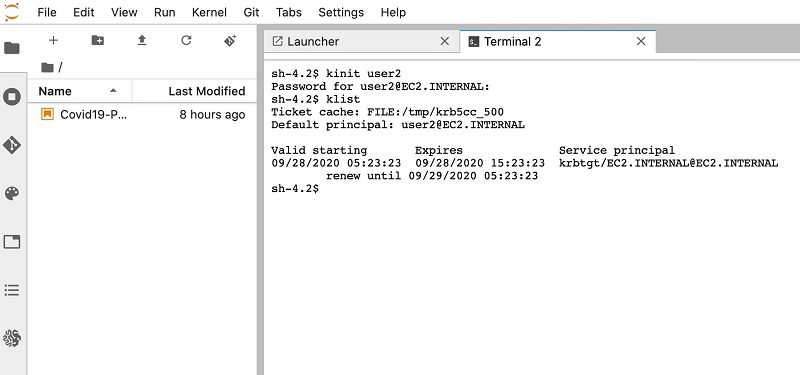
After your user is authenticated, they can access the resources associated with the IAM role defined in Amazon EMR.
Running the example notebook
To run your notebook, complete the following steps:
- Choose the
Covid19-Pandas-Sparkexample notebook.
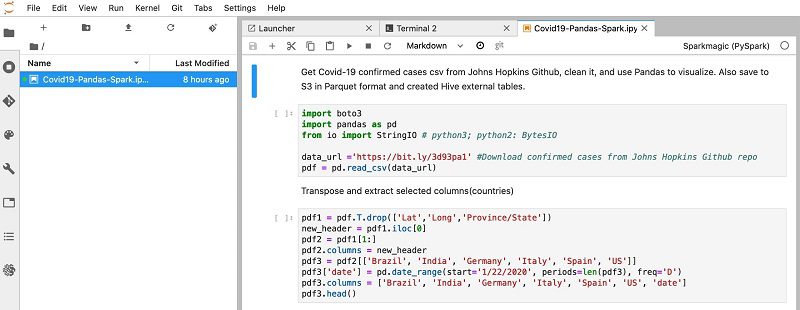
- Choose the Run (
 ) icon to progressively run the cells in the example notebook.
) icon to progressively run the cells in the example notebook.
When you reach the cell in the notebook to save the Spark DataFrame (sdf) to an internal hive table, you get an Access Denied error.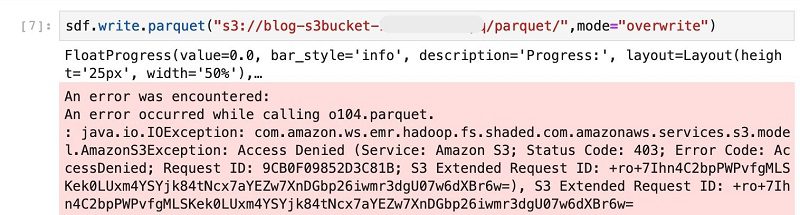
This step fails because the IAM role associated with Amazon EMR user2 doesn’t have permissions to write to the S3 bucket blog-s3bucket-xxxxx.
- Navigate back to the Terminal
- Enter
kinitfollowed by the usernameuser1. - Enter the user’s password.
The initial default password is pwd1. The first time logging in, you’re prompted to change the password.
- Enter a new password.
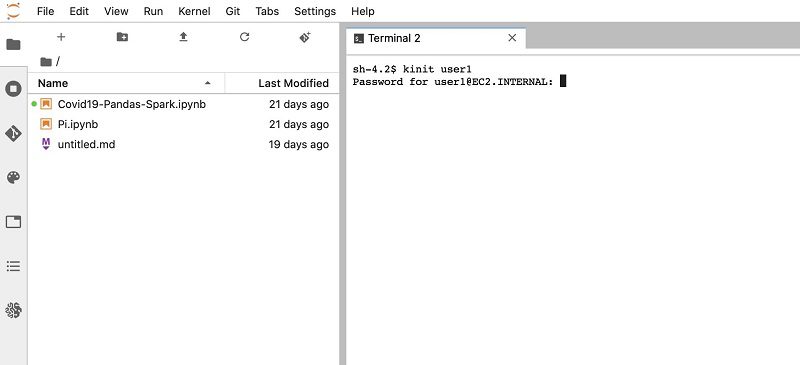
- Restart the kernel.
- Run all cells in the notebook by choosing Kernel and choosing Restart Kernel and Run All Cells.
This re-establishes a new Livy session with the EMR cluster using the new Kerberos token for user1.
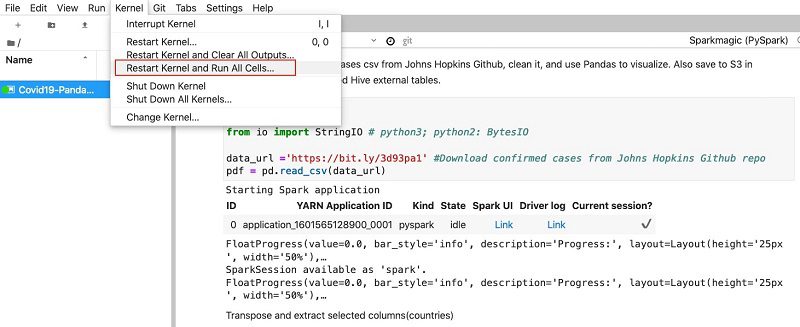
The notebook uses Pandas and Matplotlib to process and transform the raw COVID-19 dataset into a consumable format and visualize it.
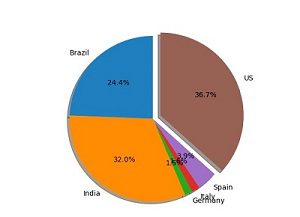
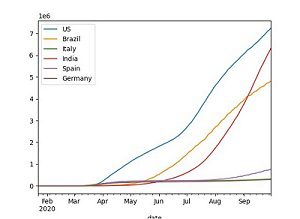
The notebook also demonstrates the creation of a native Hive table in HDFS and an external table hosted on Amazon S3, which are queried by SparkSQL. The notebook is self-explanatory; you can follow the steps to complete the demo.
Restricting principal access to Amazon S3 resources in the IAM role
The CloudFormation template by default allows any principal to assume the role blog-allowEMRFSAccessForUser1. This is apparently too permissive. We need to further restrict the principals that can assume the role.
- On the IAM console, under Access management, choose Roles.
- Search for and choose the role
blog-allowEMRFSAccessForUser1.
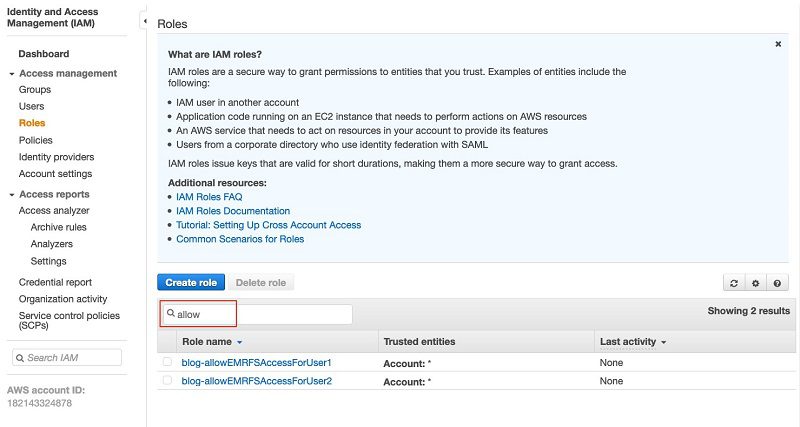
- On the Trust relationship tab, choose Edit trust relationship.
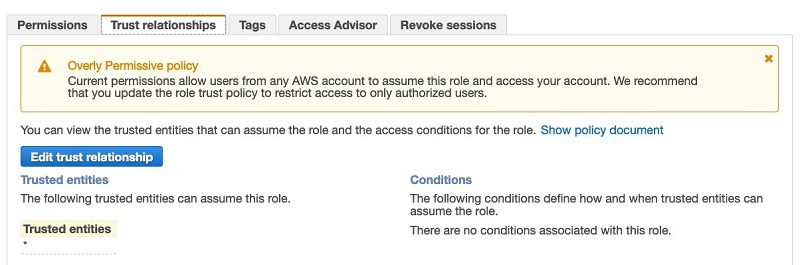
- Open a second browser window to look up your EMR cluster’s instance profile role name.
You can find the instance profile name on the IAM console by searching for the keyword instanceProfileRole. Typically, the name looks like
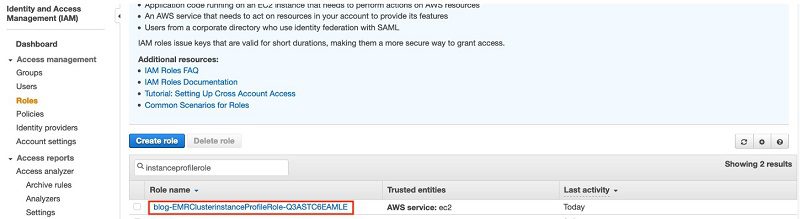
- Modify the policy document using the following JSON file, providing your own AWS account ID and the instance profile role name:
- Return to the first browser window.
- Choose Update Trust Policy.
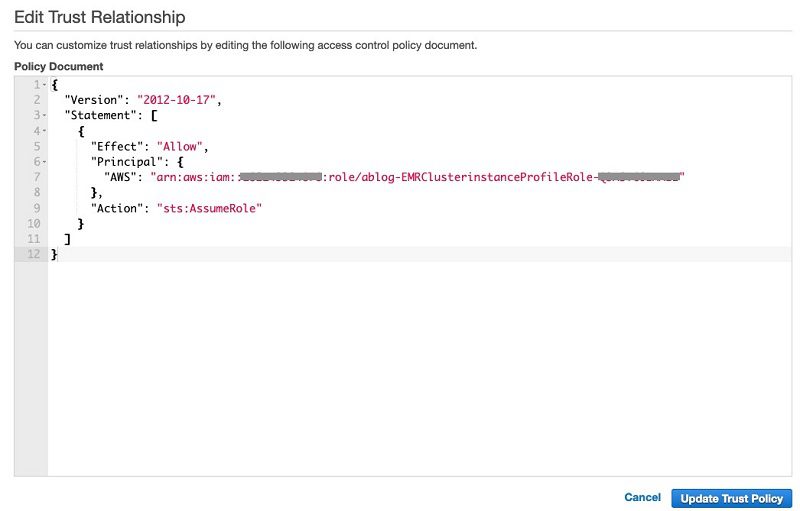
This makes sure that only the EMR cluster’s users are allowed to access their own S3 buckets.
Cleaning up
You can complete the following steps to clean up resources deployed for this solution. This also deletes the S3 bucket, so you should copy the contents in the bucket to a backup location if you want to retain the data for later use.
- On the CloudFormation console, choose Stacks.
- Select the slack deployed for this solution.
- Choose Delete.
Summary
We walked through the solution using a SageMaker notebook instance authenticated with a Kerberized EMR cluster via Apache Livy, and processed a public COVID-19 dataset with Pandas before saving it in Parquet format in an external Hive table. The table references the data hosted in an Amazon S3 bucket. We provided a CloudFormation template to automate the deployment of necessary AWS services for the demo. We strongly encourage you to use these managed and serverless services such as Amazon Athena and Amazon QuickSight for your specific use cases in production.
About the Authors
 James Sun is a Senior Solutions Architect with Amazon Web Services. James has over 15 years of experience in information technology. Prior to AWS, he held several senior technical positions at MapR, HP, NetApp, Yahoo, and EMC. He holds a PhD from Stanford University.
James Sun is a Senior Solutions Architect with Amazon Web Services. James has over 15 years of experience in information technology. Prior to AWS, he held several senior technical positions at MapR, HP, NetApp, Yahoo, and EMC. He holds a PhD from Stanford University.
 Graham Zulauf is a Senior Solutions Architect. Graham is focused on helping AWS’ strategic customers solve important problems at scale.
Graham Zulauf is a Senior Solutions Architect. Graham is focused on helping AWS’ strategic customers solve important problems at scale.
Tags: Archive

 ) icon to progressively run the cells in the example notebook.
) icon to progressively run the cells in the example notebook.
Leave a Reply