Fine-tune and deploy the ProtBERT model for protein classification using Amazon SageMaker
Proteins, the key fundamental macromolecules governing in biological bodies, are composed of amino acids. These 20 essential amino acids, each represented by a capital letter, combine to form a protein sequence, which can be used to predict the subcellular localization (the location of protein in a cell) and structure of proteins.
Figure 1: Protein Sequence
The study of protein localization is important to comprehend the function of protein, which is essentially to structure, function, and regulate the body’s tissues and organs. Protein localization has great importance for drug design and other applications. For example, we can investigate methods to disrupt the binding of the spiky S1 protein of the SARS-Cov-2 virus. The binding of the S1 protein to the human receptor ACE2 is the mechanism which led to the COVID-19 pandemic [1]. It also plays an important role in characterizing the cellular function of hypothetical and newly discovered proteins [2].
Figure 2: SARS-Cov-2 virus binding to ACE2 human receptor
Protein sequences are constrained to adopting particular 3D shapes (referred to as protein 3D structure) optimized for accomplishing particular functions. These constraints mirror the rules of grammar and meaning in natural language, thereby allowing us to map algorithms from natural language processing (NLP) directly onto protein sequences. During training, the language model learns to extract those constraints from millions of examples and store the derived knowledge in its weights. [1] Although existing solutions in protein bioinformatics [11, 12, 13, 14, 15,16] usually have to search for evolutionary-related proteins in exponentially growing databases, language models offer a potential alternative to this increasingly time-consuming database search because they extract features directly from single protein sequences. Additionally, the performance of existing solutions deteriorates if a sufficient number of related sequences can’t be found; for example, the quality of predicted protein structures correlates strongly with the number of effective sequences found in today’s databases [17].
Several research endeavors currently aim to localize whole proteomes by using high-throughput approaches [2, 3, 4]. These large datasets provide important information about protein function, and more generally global cellular processes. However, they currently don’t achieve 100% coverage of proteomes, and the methodology used can in some cases cause mislocalization of subsets of proteins [5, 6]. Therefore, complementary methods are necessary to address these problems.
In this post, we use NLP techniques for protein sequence classification. The idea is to interpret protein sequences as sentences and their constituent—amino acids—as single words [7]. More specifically, we fine-tune the PyTorch ProtBERT model from the Hugging Face library using Amazon SageMaker.
What is ProtBERT?
ProtBERT is a pretrained model on protein sequences using a masked language modeling objective. It’s based on the BERT model, which is pretrained on a large corpus of protein sequences in a self-supervised fashion. This means it was pretrained on the raw protein sequences only, with no humans labeling them in any way (which is why it can use lots of publicly available data) with an automatic process to generate inputs and labels from those protein sequences [8]. For more information about ProtBERT, see ProtTrans: Towards Cracking the Language of Life’s Code Through Self-Supervised Deep Learning and High Performance Computing.
Solution overview
The post focuses on fine-tuning the PyTorch ProtBERT model (see the following diagram). We first extend the pretrained ProtBERT model to classify the protein sequences.
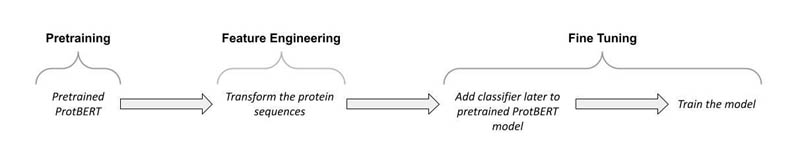
We then deploy the model using SageMaker, which is the most comprehensive and fully managed machine learning (ML) service. With SageMaker, data scientists and developers can quickly and easily build and train ML models, and then directly deploy them into a production-ready hosted environment. During the training, we use the distributed data parallel (SDP) feature in SageMaker, which extends its training capabilities on deep learning models with near-linear scaling efficiency, achieving fast time-to-train with minimal code changes.
The notebook and code from this post are available on GitHub. To run it yourself, clone the GitHub repository and open the Jupyter notebook file.
Dataset
In this post, we use an open-source DeepLoc [10] public dataset of protein sequences to train the model. The dataset is a FASTA file composed of header and protein sequence. The header is composed of the accession number from Uniprot, the annotated subcellular localization, and possibly a description field indicating if the protein was part of the test set. The subcellular localization includes an additional label, where S indicates soluble, M membrane, and U unknown [9]. The following code is a sample of the data:
A sequence in FASTA format begins with a single-line description, followed by lines of sequence data. The definition line (defline) is distinguished from the sequence data by a greater-than (>) symbol at the beginning. The word following the > symbol is the identifier of the sequence, and the rest of the line is the description.
We download the FASTA formatted dataset and read it by directly filtering out the columns that are of interest. The dataset consists of 14,000 sequences and 6 columns in total. The columns are as follows:
- id – Unique identifier given each sequence in the dataset.
- sequence – Protein sequence. Each character is separated by a space. This is useful for the BERT tokenizer.
- sequence_length – Character length of each protein sequence.
- location – Classification given each sequence. The dataset has 10 unique classes (subcellular localization).
- is_train – Indicates whether the record should be used for training or test. Is also used to separate the dataset for training and validation.
When we plot the sequence lengths of each record as an histogram, we observe the following distribution.
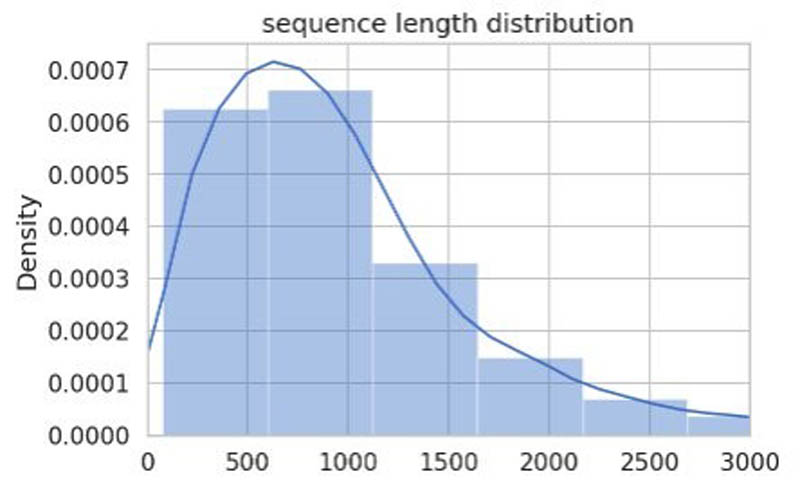
This is an important observation because the ProtBERT model receives a fixed sentence length as input. Usually, the maximum length of a sentence depends on the data we’re working on. For sentences that are shorter than this maximum length, we have to add paddings (empty tokens) to the sentences to make up the length.
In the preceding plot, most of the sequences are under 1,500 characters in length, therefore, it’s a good idea to choose max_length = 1536, but that increases the training time for this sample notebook, therefore, we use max_length = 512.
When we’re retrieving each sequence record using the Pytorch DataLoaders during training, we must ensure that each sequence is tokenized, truncated, and the necessary padding is added to make them all the same max_length value. To encapsulate this process, we define the ProteinSequenceDataset class, which uses the encode_plus() API provided by the Hugging Face transformer library:
Next, we divide the dataset into training and test. We can use the is_train column to do the split, which results 11,231 records for the training set and 2,773 records for the test set (about a 75:25 data split). Finally, we upload this test and train data to our Amazon Simple Storage Service (Amazon S3) location in order to accommodate model training on SageMaker.
ProtBERT fine-tuning
In computational biology and bioinformatics, we have gold mines of data from protein sequences, but we need high computing resources to train the models, which can be limiting and costly. One way to overcome this challenge is to use transfer learning.
Transfer learning is an ML method in which a pretrained model, such as a pretrained BERT model for text classification, is reused as the starting point for a different but related problem. By reusing parameters from pretrained models, you can save significant amounts of training time and cost.
In our notebook, we use the pretrained prot_bert_bfd_localization model on the DeepLoc dataset for predicting protein subcellular localization by adding a classification layer, as shown in the following code:
We use ProteinClassifier defined in the model_def.py script for training.
Training script
We use the PyTorch-Transformers library, which contains PyTorch implementations and pretrained model weights for many NLP models, including BERT. As mentioned earlier, we use the ProtBERT model, which is pretrained on protein sequences.
We also use the distributed data parallel feature launched in December 2020 to speed up the training by distributing the data on multiple GPUs. It’s very similar to a PyTorch training script you might run outside of SageMaker, but modified to run with SDP. SDP’s PyTorch client provides an alternative to PyTorch’s native DDP. For details about how to use SDP in your native PyTorch script, see the Get Started with Distributed Training.
The following script saves the model artifacts learned during training to a file path, model_dir, as mandated by the SageMaker PyTorch image:
When training is complete, SageMaker uploads model artifacts saved in model_dir to Amazon S3 so they’re available for deployment. The following code in the script saves the trained model artifacts:
Because PyTorch-Transformer isn’t included natively in SageMaker PyTorch images, we have to provide a requirements.txt file so that SageMaker installs this library for training and inference. A requirements.txt file is a text file that contains a list of items that are installed by using pip install. You can also specify the version of an item to install. To install PyTorch-Transformer and other libraries, we add the following line to the requirements.txt file:
You can view the entire file in the GitHub repo, and it also goes into the code/ directory. For more information about the format of a requirements.txt file, see Requirements Files.
Train on SageMaker
We use SageMaker to train and deploy a model using our custom PyTorch code. The SageMaker Python SDK makes it easy to run a PyTorch script in SageMaker using its PyTorch estimator. After that, we can use the SageMaker Python SDK to deploy the trained model and run predictions. For more information on how to use this SDK with PyTorch, see Use PyTorch with the SageMaker Python SDK.
To start, we use the PyTorch estimator class to train our model. When creating our estimator, we make sure to specify a few things:
- entry_point – The name of our PyTorch script. It contains our training script, which loads data from the input channels, configures training with hyperparameters, trains a model, and saves the model. It also contains code to load and run the model during inference.
- source_dir – The location of our training scripts and
requirements.txtfile. The requirements file lists packages you want to use with your script. - framework_version – The PyTorch version we want to use.
The PyTorch estimator supports both single-machine and multi-machine, distributed PyTorch training using SDP. Our training script supports distributed training for only GPU instances.
Instance types
SDP supports model training on SageMaker with the following instance types only:
- p3.16xlarge
- p3dn.24xlarge (Recommended)
- p4d.24xlarge (Recommended)
Instance count
To get the best performance out of SDP, you should use at least two instances, but you can also use one for testing this example, which implements the script in a single instance, multiple GPU mode, taking advantage of the eight GPUs on the instance to train faster and cheaper.
Distribution strategy
To use DDP mode, you update the the distribution strategy and set it to use smdistributed dataparallel.
After we create the estimator, we call fit(), which launches a training job. We use the Amazon S3 URIs that we uploaded the training data to earlier. See the following code:
With max_length=512 and running the model for only three epochs, we get a validation accuracy of around 65%, which is pretty decent. You can optimize it further by trying a bigger sequence length, increasing the number of epochs, and tuning other hyperparameters. Make sure to increase the GPU memory or reduce the batch size when you increase the sequence length, otherwise you might get cuda out of memory error.
For more details on optimizing the model, see ProtTrans: Towards Cracking the Language of Life’s Code Through Self-Supervised Deep Learning and High Performance Computing.
Deploy the model on SageMaker
After we train our model, we host it on an SageMaker endpoint. To make the endpoint load the model and serve predictions, we implement a few methods in inference.py:
- model_fn() – Loads the saved model and returns a model object that can be used for model serving. The SageMaker PyTorch model server loads our model by invoking
model_fn:
- input_fn() – Deserializes and prepares the prediction input. In this example, our request body is first serialized to JSON and then sent to the model serving endpoint. Therefore, in
input_fn(), we first deserialize the JSON-formatted request body and return the input as atorch.tensor, as required for the ProtBERT model:
- predict_fn() – Performs the prediction and returns the result. To deploy our endpoint, we call
deploy()on our PyTorch estimator object, passing in our desired number of instances and instance type:
Create a model object
You define the model object by using the SageMaker SDK’s PyTorchModel and pass in the model from the estimator and the entry_point. The function loads the model and sets it to use a GPU, if available. See the following code:
Deploy the model on an endpoint
You create a predictor by using the model.deploy function. You can optionally change both the instance count and instance type:
Predict protein subcellular localization
Now that we have deployed the model endpoint, we can provide some protein sequences and let the model endpoint identify their subcellular localization, using the predictor we created:
The following table summarizes some of our results.
| Sequence | Ground Truth | Prediction |
| M G K K D A S T T R T P V D Q Y R K Q I G R Q D Y K K N K P V L K A T R L K A E A K K A A I G I K E V I L V T I A I L V L L F A F Y A F F F L N L T K T D I Y E D S N N | Endoplasmic.reticulum | Endoplasmic.reticulum |
| M S M T I L P L E L I D K C I G S N L W V I M K S E R E F A G T L V G F D D Y V N I V L K D V T E Y D T V T G V T E K H S E M L L N G N G M C M L I P G G K P E | Nucleus | Nucleus |
| M G G P T R R H Q E E G S A E C L G G P S T R A A P G P G L R D F H F T T A G P S K A D R L G D A A Q I H R E R M R P V Q C G D G S G E R V F L Q S P G S I G T L Y I R L D L N S Q R S T C C C L L N A G T K G M C | Cytoplasm | Cytoplasm |
Clean up resources
Remember to delete the SageMaker endpoint and SageMaker notebook instance created to avoid charges. See the following code:
Conclusion
In this post, we used a pretrained ProtBERT model (prot_bert_bfd_localization) as a starting point and fine-tuned it for the downstream task of identifying the subcelluar localization of protein sequences. We used the SageMaker capabilities to train, deploy, and do the inference. Furthermore, we explored the SageMaker data parallel feature to make our training process efficient. You can use the same concept to perform other downstream tasks, such as for amino-acid level classification like predicting the secondary structure of the protein. For more about using PyTorch with SageMaker, see Using PyTorch with the SageMaker Python SDK.
References
- [1] ProtTrans: Towards Cracking the Language of Life’s Code Through Self-Supervised Deep Learning and High Performance Computing (https://www.biorxiv.org/content/10.1101/2020.07.12.199554v2.full.pdf)
- [2]Protein sequence Diagram : https://www.technologynetworks.com/applied-sciences/articles/essential-amino-acids-chart-abbreviations-and-structure-324357
- [3] Refining Protein Subcellular Localization (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1289393/)
- [4] Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. [PMC free article] [PubMed] [Google Scholar]
- [5] Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. [PubMed] [Google Scholar]
- [6] Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, et al. From ORFeome to biology: A functional genomics pipeline. Genome Res. 2004;14:2136–2144. [PMC free article] [PubMed] [Google Scholar]
- [7] Davis TN. Protein localization in proteomics. Curr Opin Chem Biol. 2004;8:49–53. [PubMed] [Google Scholar]
- [8] Scott MS, Thomas DY, Hallett MT. Predicting subcellular localization via protein motif co-occurrence. Genome Res. 2004;14:1957–1966. [PMC free article] [PubMed] [Google Scholar]
- [9] ProtBERT Hugging Face (https://huggingface.co/Rostlab/prot_bert)
- [10] DeepLoc-1.0: Eukaryotic protein subcellular localization predictor (http://www.cbs.dtu.dk/services/DeepLoc-1.0/data.php)
- [11] M. S. Klausen, M. C. Jespersen et al., “NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning,” Proteins: Structure, Function, and Bioinformatics, vol. 87, no. 6, pp. 520–527, 2019, _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.25674.
- [12] J. J. Almagro Armenteros, C. K. Sønderby et al., “DeepLoc: Prediction of protein subcellular localization using deep learning,” Bioinformatics, vol. 33, no. 21, pp. 3387–3395, Nov. 2017.
- [13] J. Yang, I. Anishchenko et al., “Improved protein structure prediction using predicted interresidue orientations,” Proceedings of the National Academy of Sciences, vol. 117, no. 3, pp. 1496–1503, Jan. 2020.
- [14] A. Kulandaisamy, J. Zaucha et al., “Pred-MutHTP: Prediction of disease-causing and neutral mutations in human transmembrane proteins,” Human Mutation, vol. 41, no. 3, pp. 581–590, 2020, _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/humu.23961.
- [15] M. Schelling, T. A. Hopf, and B. Rost, “Evolutionary couplings and sequence variation effect predict protein binding sites,” Proteins: Structure, Function, and Bioinformatics, vol. 86, no. 10, pp. 1064–1074, 2018, _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.25585.
- [16] M. Bernhofer, E. Kloppmann et al., “TMSEG: Novel prediction of transmembrane helices,” Proteins: Structure, Function, and Bioinformatics, vol. 84, no. 11, pp. 1706–1716, 2016, _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/prot.25155.
- [17] D. S. Marks, L. J. Colwell et al., “Protein 3D Structure Computed from Evolutionary Sequence Variation,” PLOS ONE, vol. 6, no. 12, p. e28766, Dec. 2011.
About the Authors
 Mani Khanuja is an Artificial Intelligence and Machine Learning Specialist SA at Amazon Web Services (AWS). She helps customers using machine learning to solve their business challenges using the AWS. She spends most of her time diving deep and teaching customers on AI/ML projects related to computer vision, natural language processing, forecasting, ML at the edge, and more. She is passionate about ML at edge, therefore, she has created her own lab with self-driving kit and prototype manufacturing production line, where she spend lot of her free time.
Mani Khanuja is an Artificial Intelligence and Machine Learning Specialist SA at Amazon Web Services (AWS). She helps customers using machine learning to solve their business challenges using the AWS. She spends most of her time diving deep and teaching customers on AI/ML projects related to computer vision, natural language processing, forecasting, ML at the edge, and more. She is passionate about ML at edge, therefore, she has created her own lab with self-driving kit and prototype manufacturing production line, where she spend lot of her free time.
 Shamika Ariyawansa is a Solutions Architect at AWS helping customers run a variety of applications on AWS and machine learning workloads in particular. He is based out of Denver, Colorado. In his spare time, he enjoys off-roading adventures in the Colorado mountains and competing in machine learning competitions.
Shamika Ariyawansa is a Solutions Architect at AWS helping customers run a variety of applications on AWS and machine learning workloads in particular. He is based out of Denver, Colorado. In his spare time, he enjoys off-roading adventures in the Colorado mountains and competing in machine learning competitions.
 Vaijayanti Joshi is a Boston-based Solutions Architect for AWS. She is passionate about technology and enjoys helping customers find innovative solutions to complex business challenges. Her core areas of focus are machine learning and analytics. When she’s not working with customers on their journey to the cloud, she enjoys biking, swimming, and exploring new places.
Vaijayanti Joshi is a Boston-based Solutions Architect for AWS. She is passionate about technology and enjoys helping customers find innovative solutions to complex business challenges. Her core areas of focus are machine learning and analytics. When she’s not working with customers on their journey to the cloud, she enjoys biking, swimming, and exploring new places.
Tags: Archive

Leave a Reply